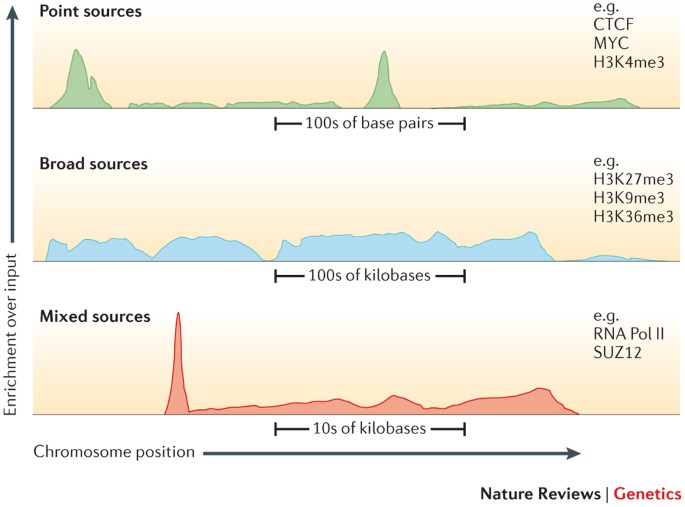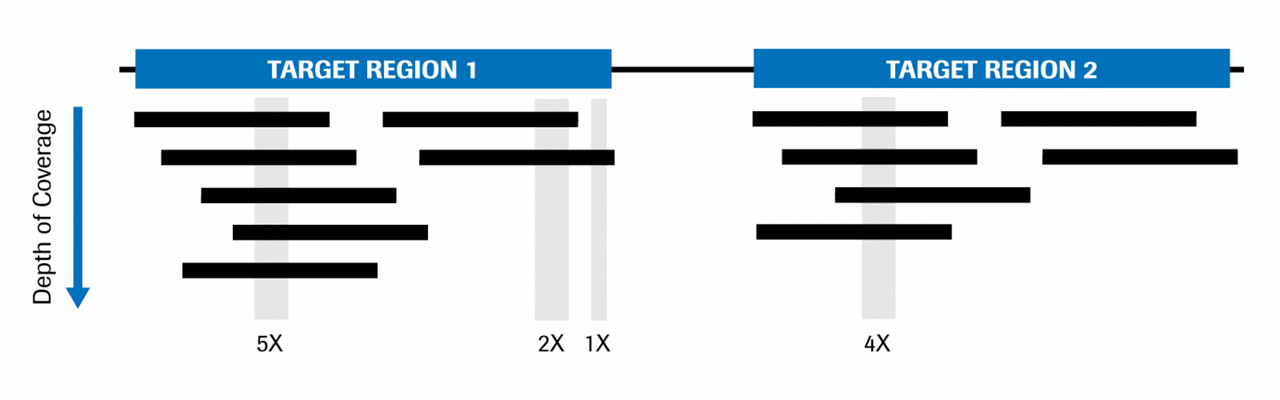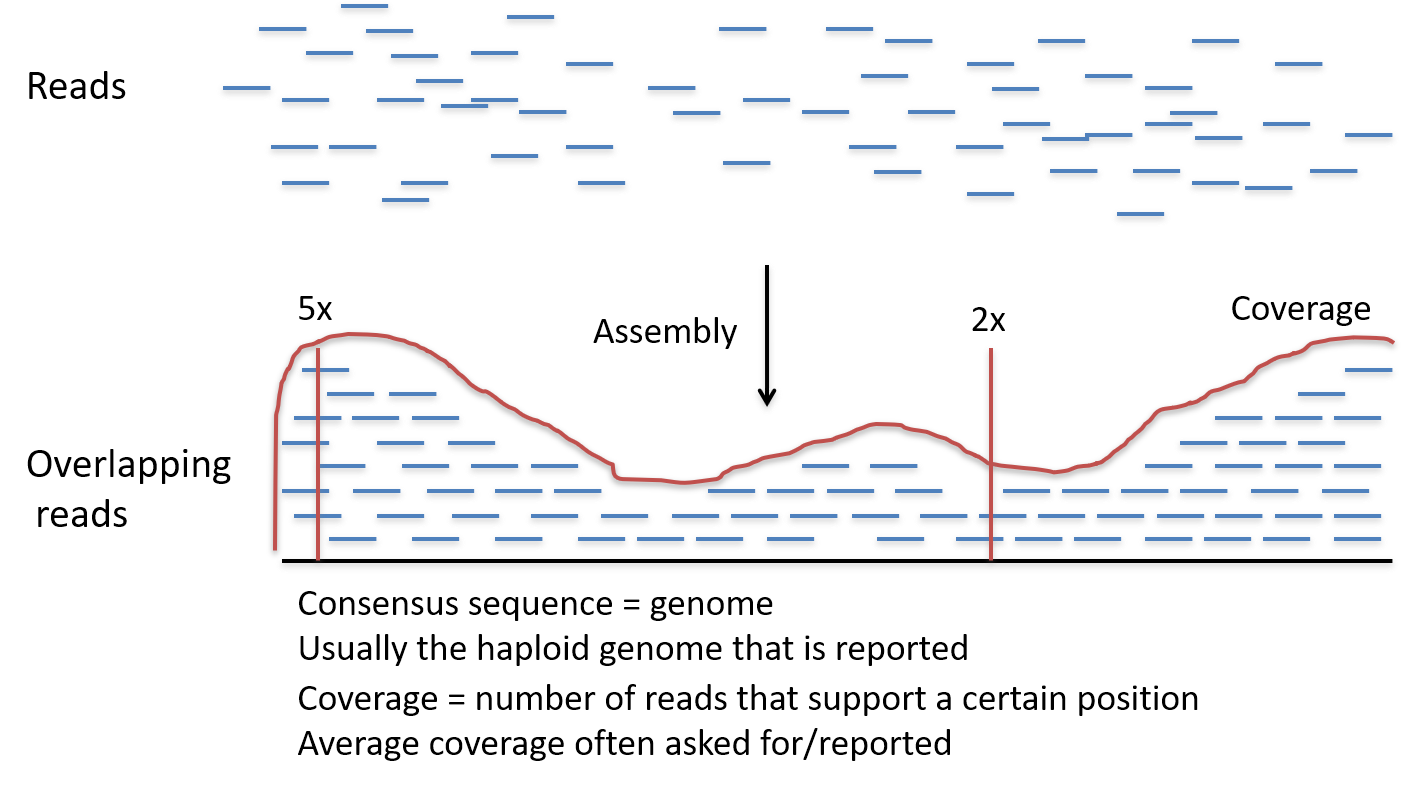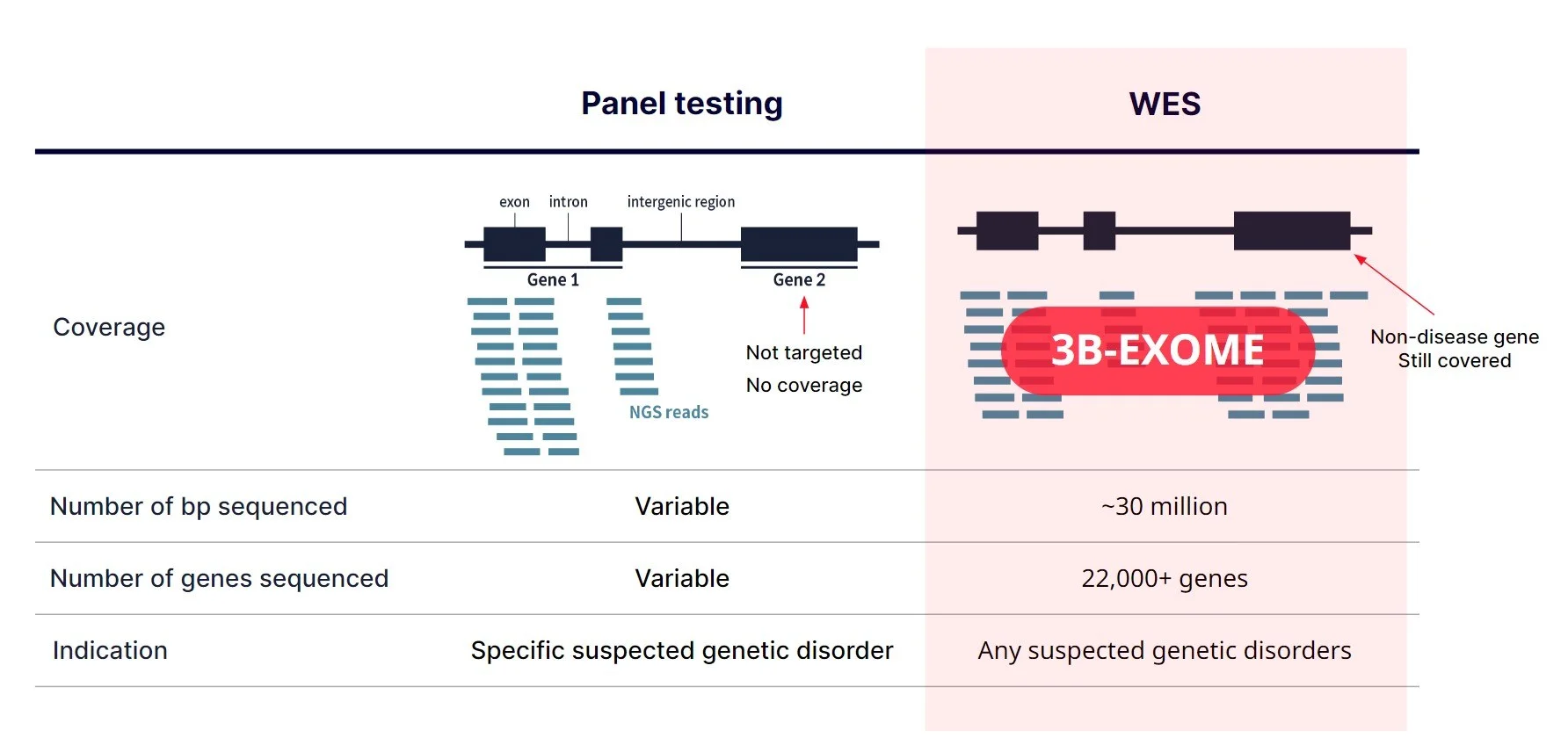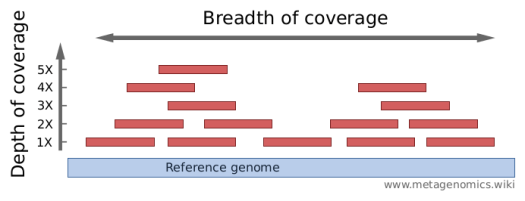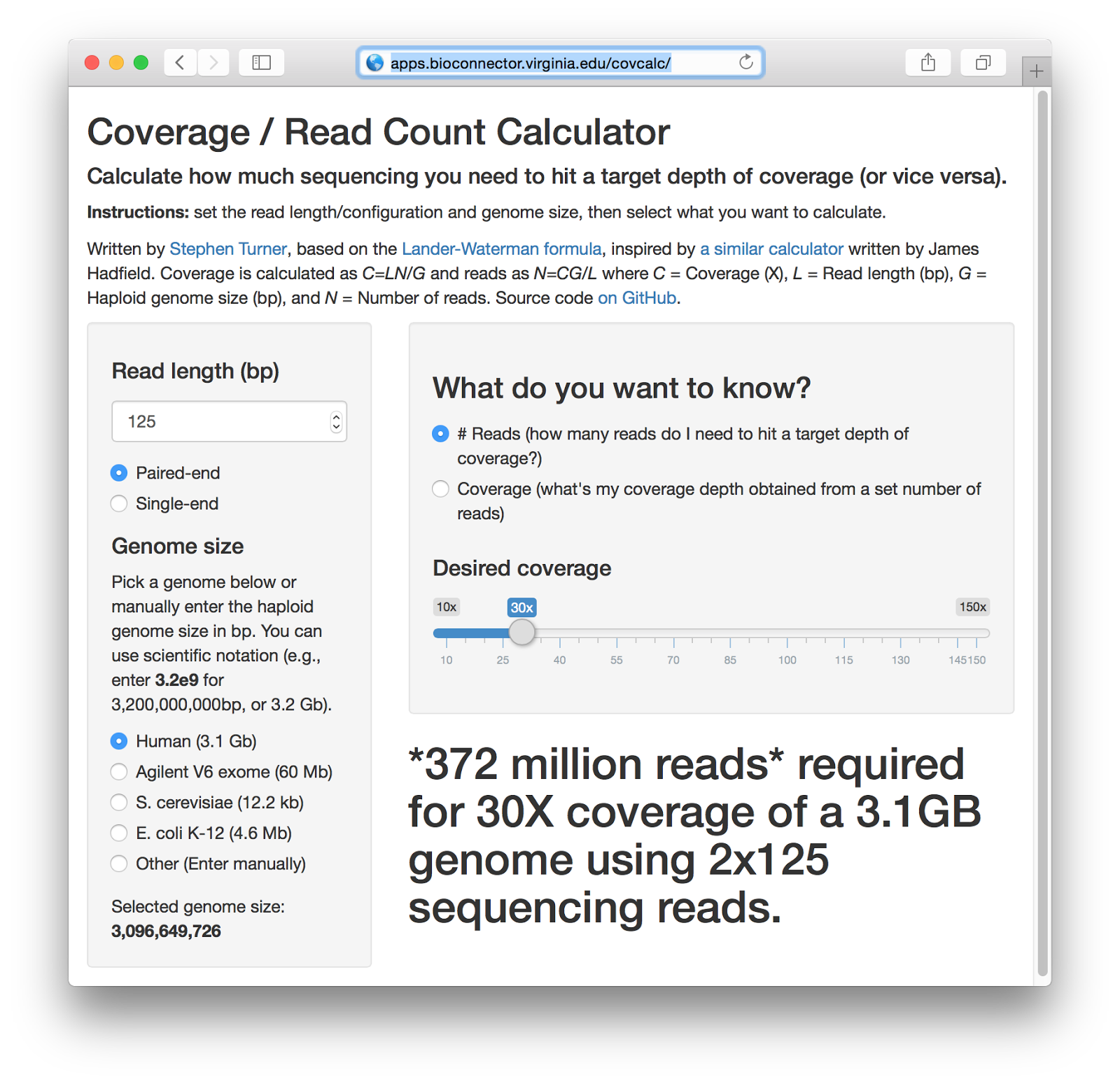
Getting Genetics Done: Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments

Detecting copy number variation in next generation sequencing data from diagnostic gene panels | BMC Medical Genomics | Full Text

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers
Sequence deeper without sequencing more: Bayesian resolution of ambiguously mapped reads | PLOS Computational Biology

Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text