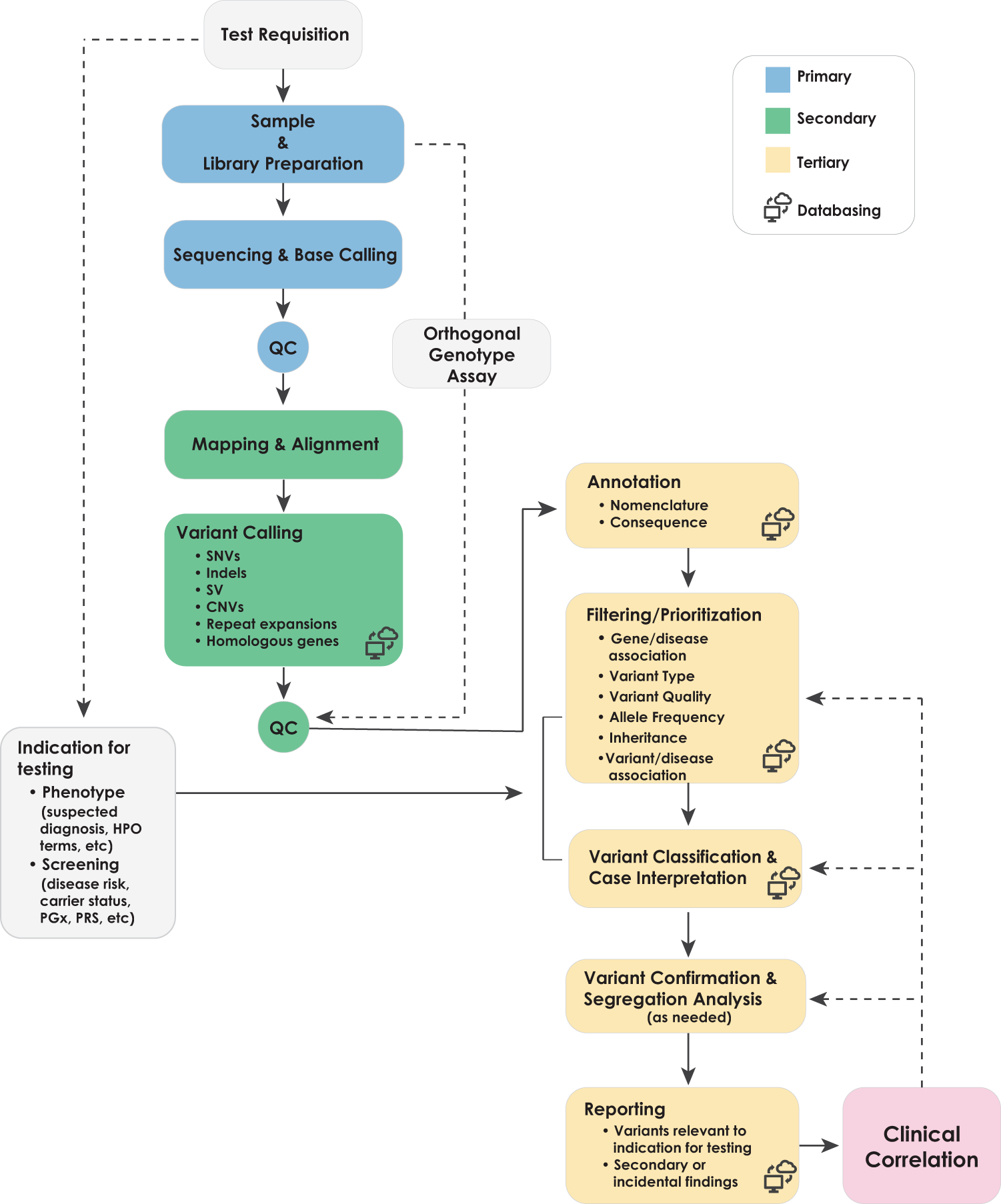
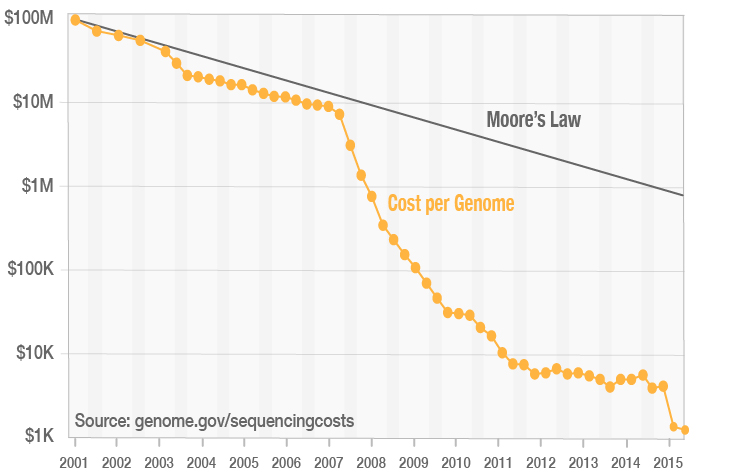
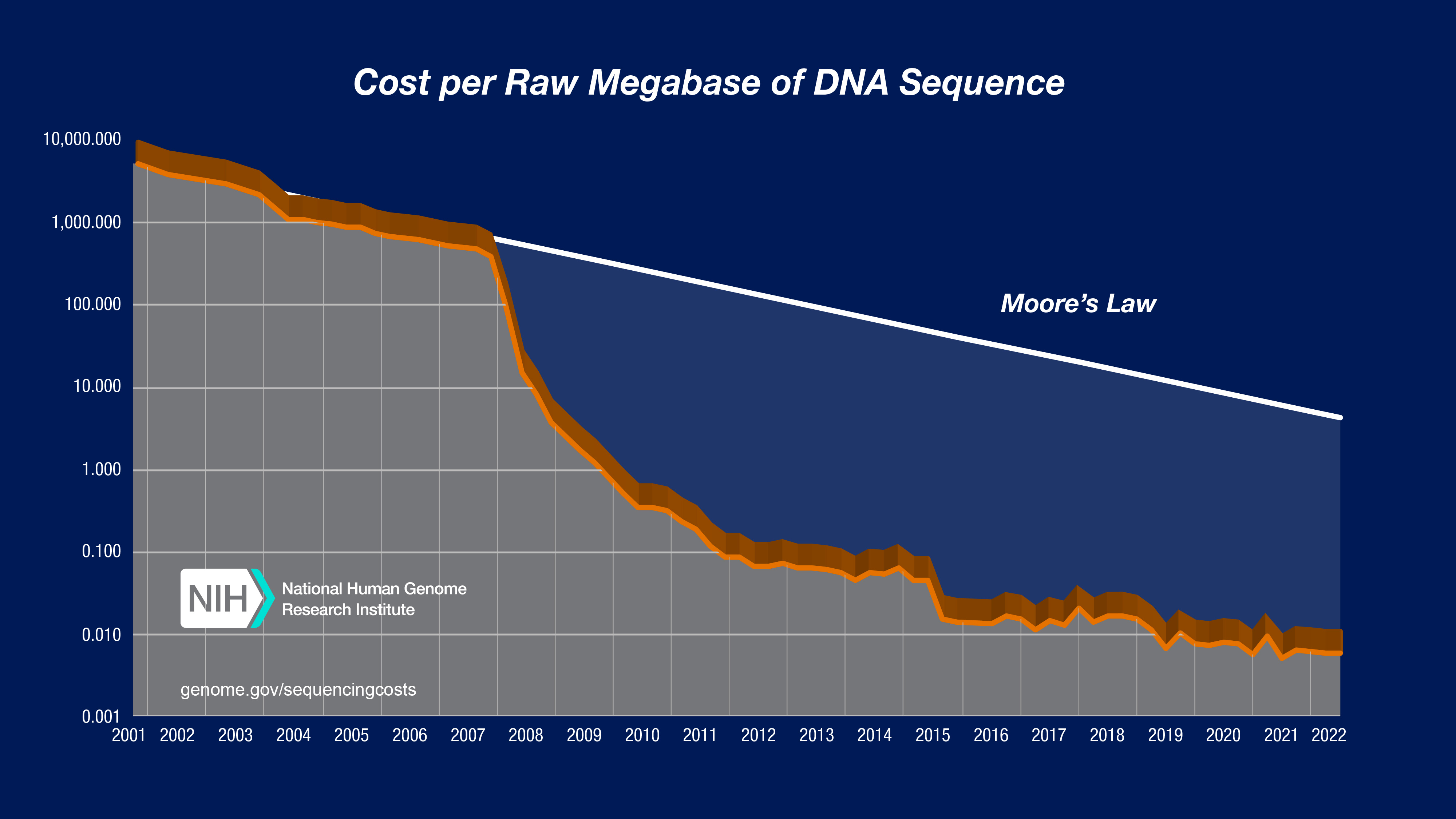
Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers
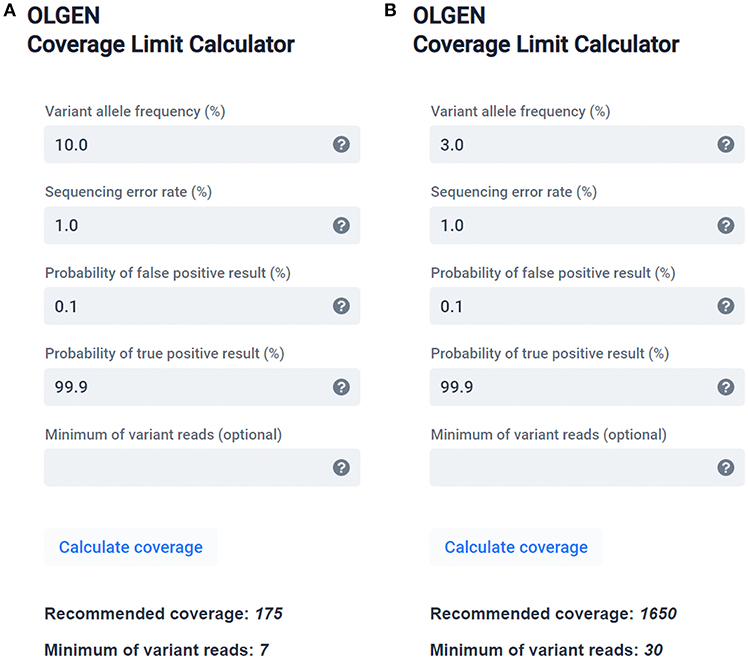
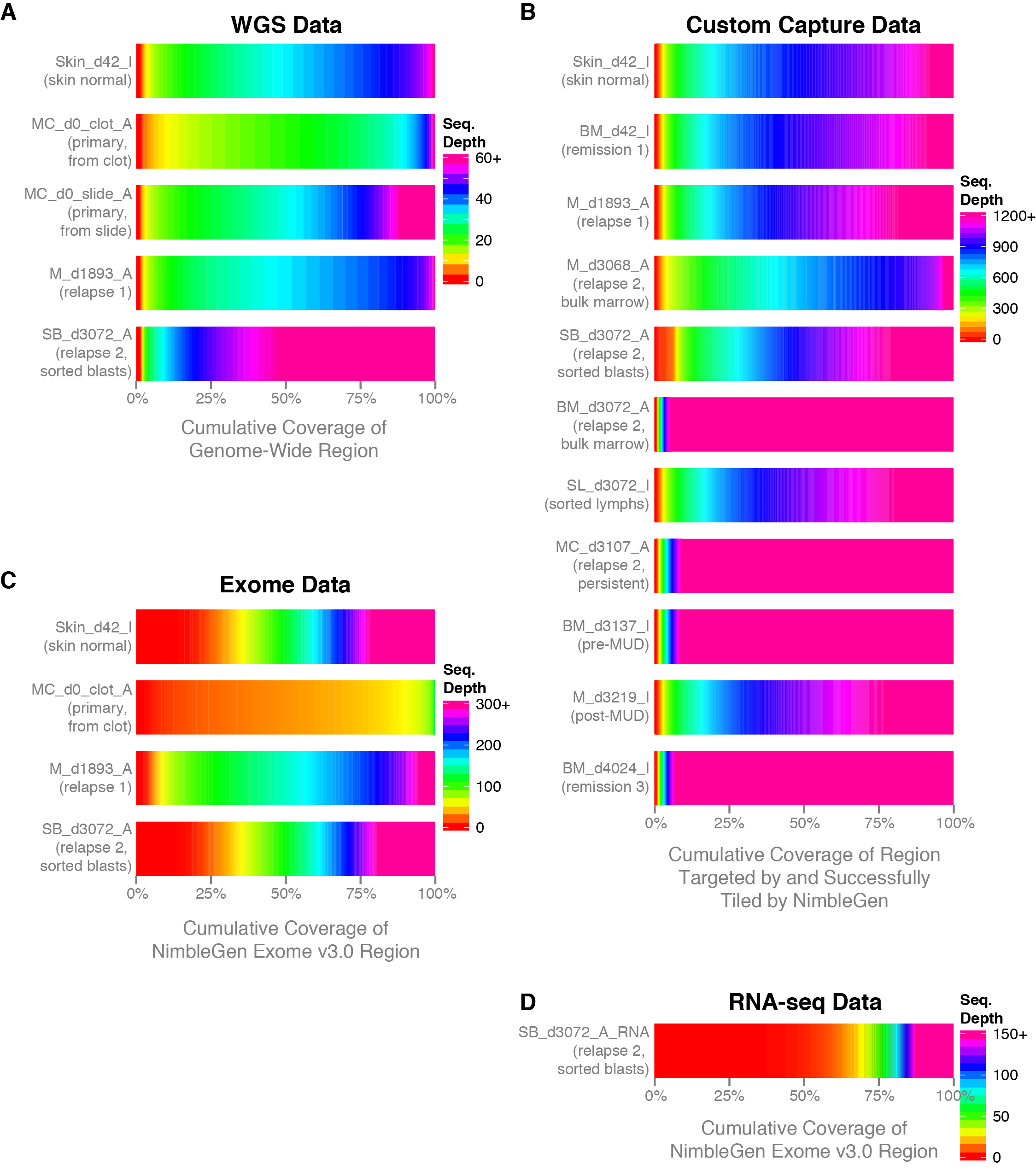
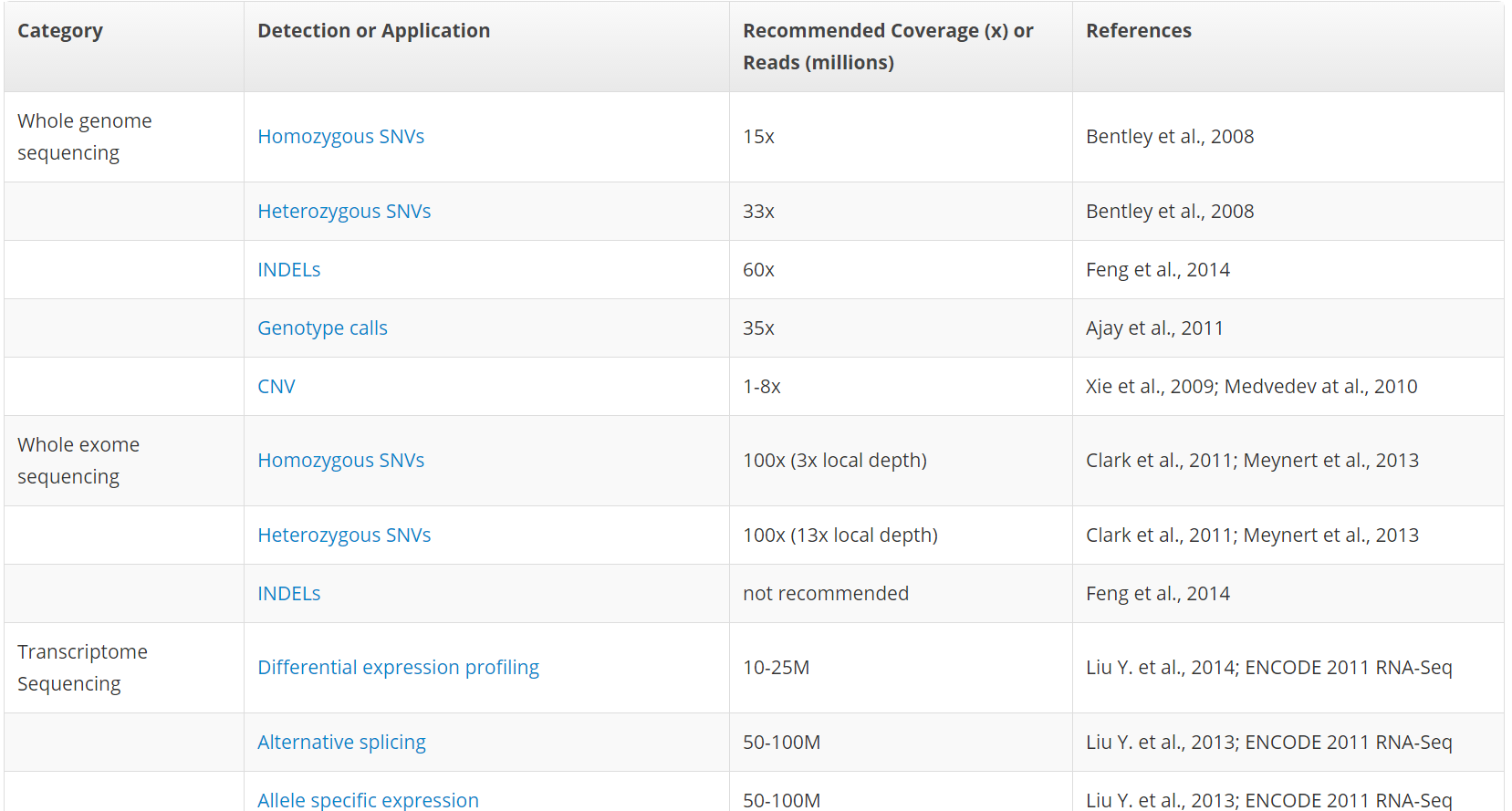
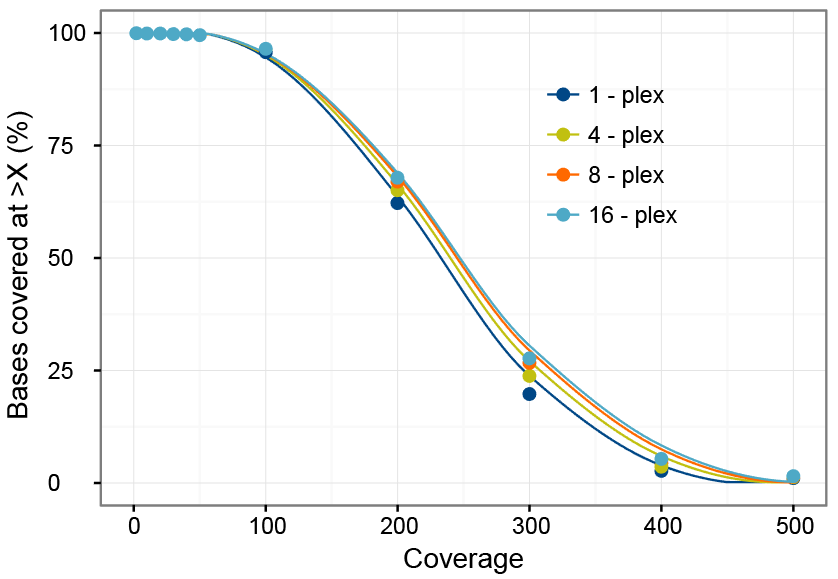
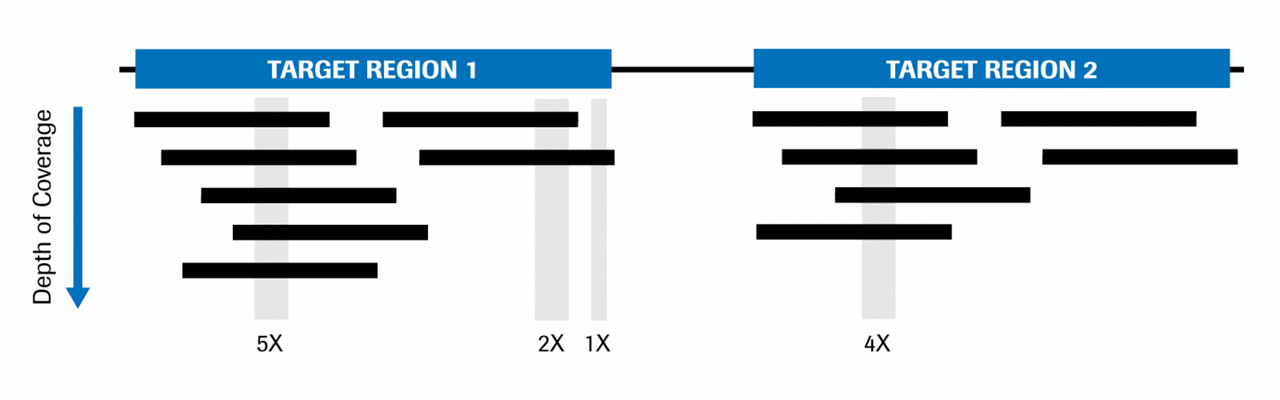
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics
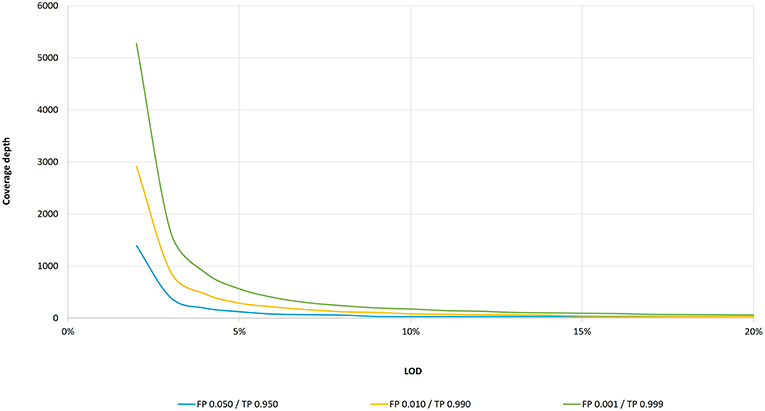
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics